Methicillin-resistant Staphylococcus aureus (MRSA) poses a serious public health threat due to its drug resistance. Characterizing metabolic pathways in MRSA may provide new insights into targeting these essential processes, potentially leading to novel strategies for controlling MRSA infections. Pearl Perl uses techniques such as Differential Radial Capillary Action of Ligand Assay (DRaCALA) and Electrophoretic Mobility Shift Assay (EMSA) to characterize the DNA-binding ability of GlnR in the presence of different regulatory proteins. In collaboration with Josh Leeming, Perl has found that the signaling protein PstA regulates the transcription factor GlnR, which in turn controls glutamine metabolism in MRSA.


Sarahi Hernandez is interested in understanding how methicillin-resistant Staphylococcus aureus (MRSA) responds to stress and survives during infection. In collaboration with Oscar Villasana Espinosa, Sarahi and Oscar have identified several genes that are important for intracellular survival during macrophage infection. Sarahi’s follow-up work is investigating how these genes regulate MRSA survival during macrophage phagocytosis, which will provide insights into MRSA pathogenesis and persistent infection.

To celebrate the publication of our first paper in mBio and the holiday season, the Tang Lab held a potluck featuring traditional foods from six different countries.
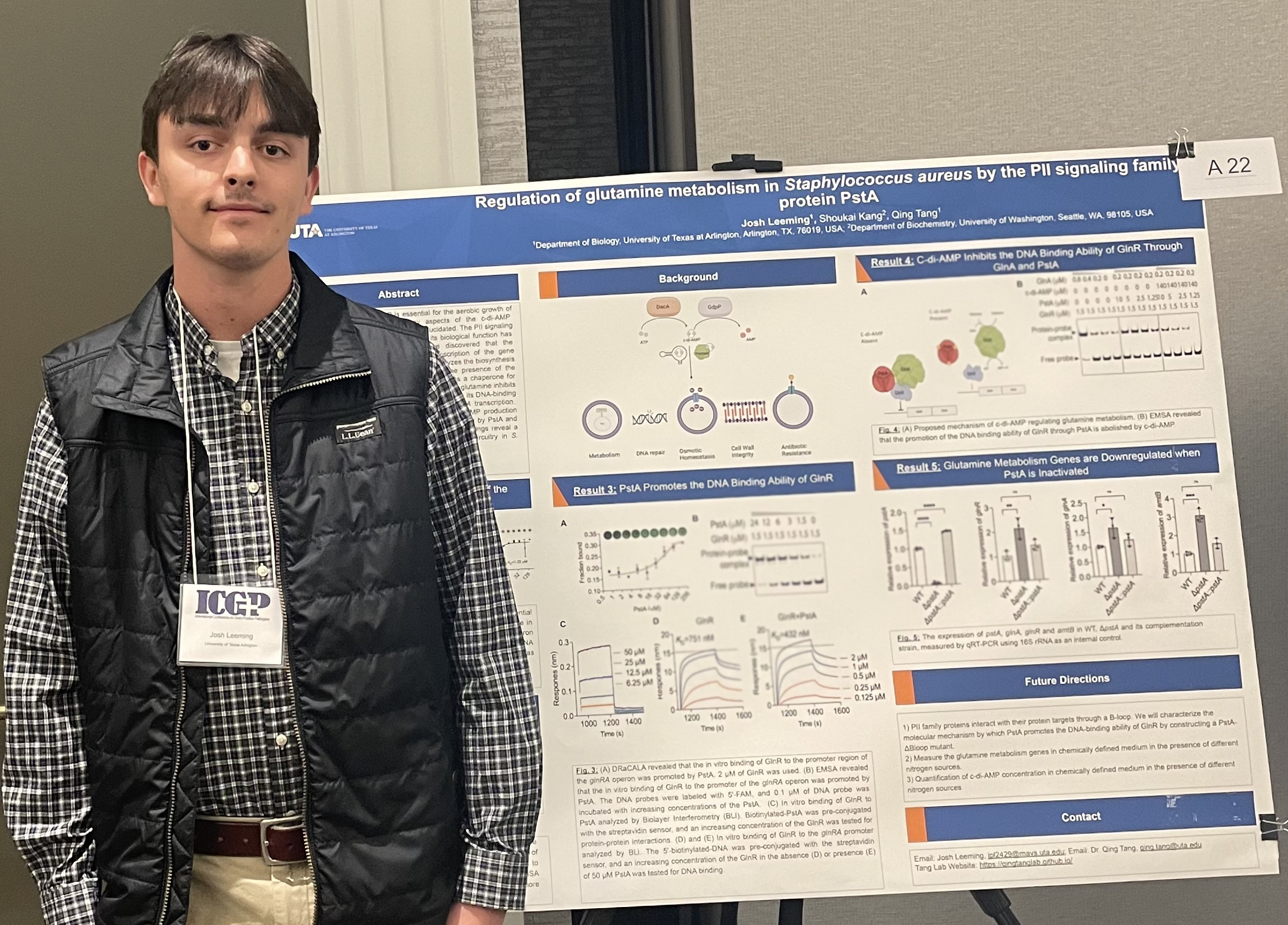
The second messenger c-di-AMP is essential for aerobic growth in Staphylococcus aureus; however, many aspects of the c-di-AMP signaling network in S. aureus remain to be elucidated. Josh’s research focuses on uncovering the regulatory role of c-di-AMP in glutamine metabolism, a process crucial for bacterial growth. He has identified a novel regulator, PstA, a PII family protein that modulates the transcription of glutamine synthesis genes. These findings reveal a previously unknown aspect of the nitrogen regulatory circuitry in S. aureus.
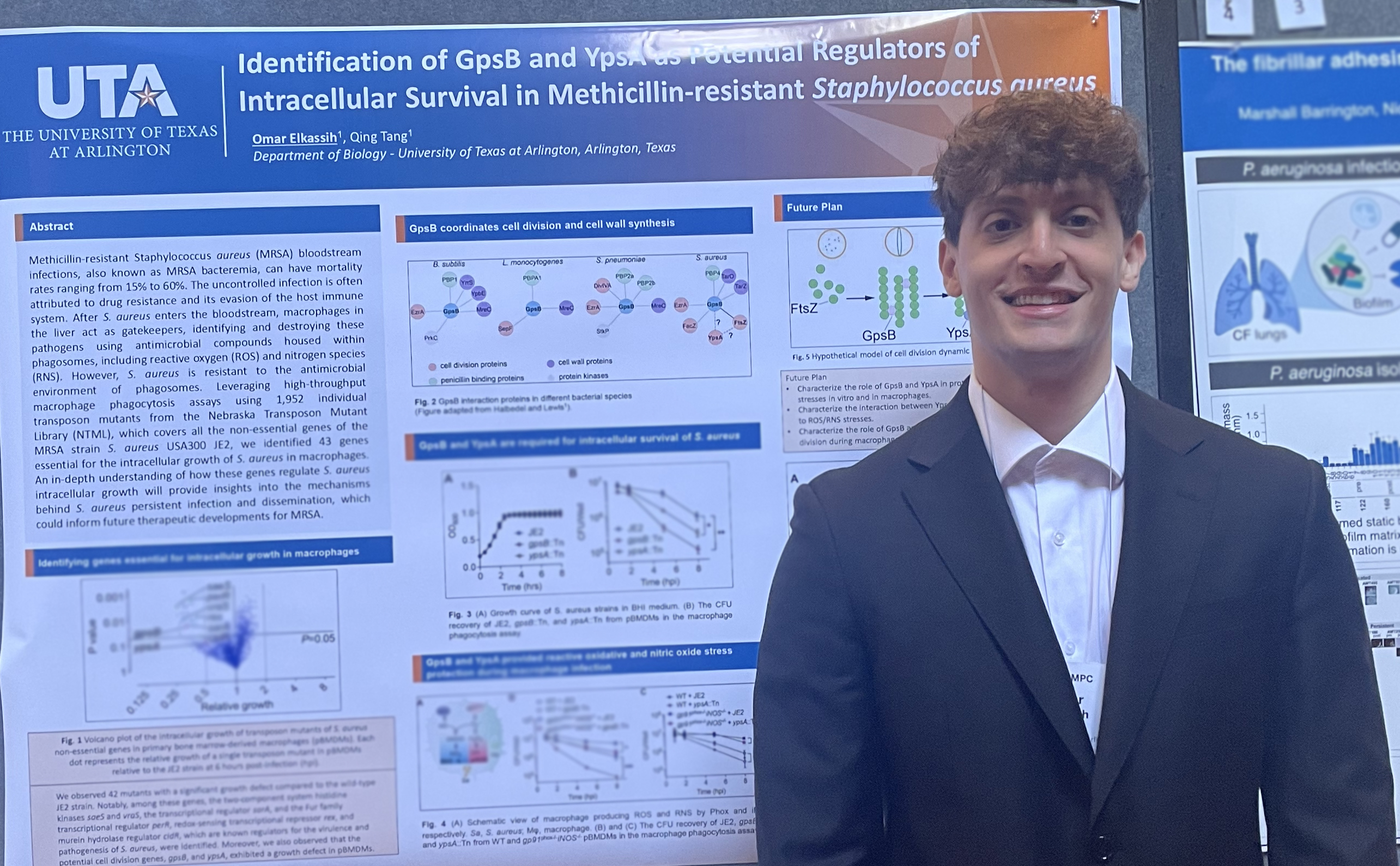
Macrophages act as the gatekeepers to detect and resolve bacterial infections. Bacteria phagocytosis by macrophages may lead to intracellular killing, destroying the bacteria by producing antimicrobials including reactive oxygen species (ROS) and nitrogen species (RNS) in the phagosomes. However, S. aureus is resistant to the antimicrobial environment of phagosomes, using these compartments to hide from immune cells and antibiotics. Ultimately, S. aureus will proliferate in the cytoplasm, and cause macrophage death, eventually enabling further bacterial dissemination. The interactions between macrophages and S. aureus directly impact the outcome of the infection. In depth understanding of these host-pathogen interactions could provide insights for future therapeutic developments for MRSA. Omar works on the identification of S. aureus genes that are essential for intracellular growth in macrophages. He has found that the potential cell division protein GpsB is not required for growth in rich medium but is necessary for intracellular growth of S. aureus in macrophages. He later found that GpsB knockout mutant is more susceptible to the ROS produced by macrophages. Omar is currently characterizing the mechanism of GpsB in pathogenesis.

Minh is an undergraduate LSAMP scholar in 2024 who spent the summer working with the Tang Lab. During her time with us, she focused on identifying Staphylococcus aureus genes essential for systemic infection in mice and made remarkable progress in just two months. Minh, you will be greatly missed by all of us.
Qing Tang officially joined Department of Biology at the University of Texas at Arlington as an assitant professor starting in January 2024!
Qing Tang’s publication, titled “Thymidine starvation promotes c-di-AMP-dependent inflammation during pathogenic bacterial infection,” has been selected as a Highlighted Article by Science Signaling.
Abstract: Bacterial stress caused by antibiotics increases a second messenger that directly stimulates host inflammation.