Abstract
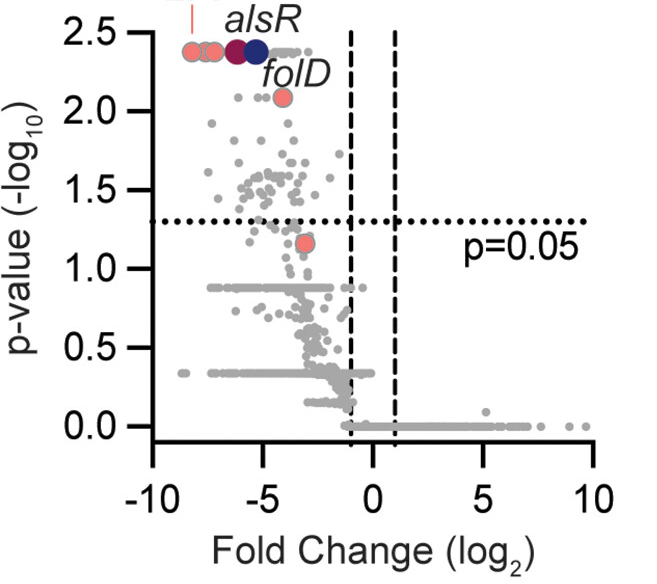
Transposon sequencing (Tn-seq) is a powerful genome-wide technique to assess bacterial fitness under varying growth conditions. However, screening via Tn-seq in vivo is challenging. Dose limitations and host restrictions create bottlenecks that diminish the transposon mutant pool being screened. Here, we have developed a murine model with a disruption in Akr1c13 that renders the resulting RECON−/− mouse resistant to high-dose infection. We leveraged this model to perform a Tn-seq screen of the human pathogen Listeria monocytogenes in vivo. We identified 135 genes which were required for L. monocytogenes growth in mice including novel genes not previously identified for host survival. We identified organ-specific requirements for L. monocytogenes survival and investigated the role of the folate enzyme FolD in L. monocytogenes liver pathogenesis. A mutant lacking folD was impaired for growth in murine livers by 2.5-log10 compared to wild type and failed to spread cell-to-cell in fibroblasts. In contrast, a mutant in alsR, which encodes a transcription factor that represses an operon involved in D-allose catabolism, was attenuated in both livers and spleens of mice by 4-log10 and 3-log10, respectively, but showed modest phenotypes in in vitro models. We confirmed that dysregulation of the D-allose catabolism operon is responsible for the in vivo growth defect, as deletion of the operon in the ∆alsR background rescued virulence. By undertaking an unbiased, genome-wide screen in mice, we have identified novel fitness determinants for L. monocytogenes host infection, which highlights the utility of the RECON−/− mouse model for future screening efforts.